|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|

|
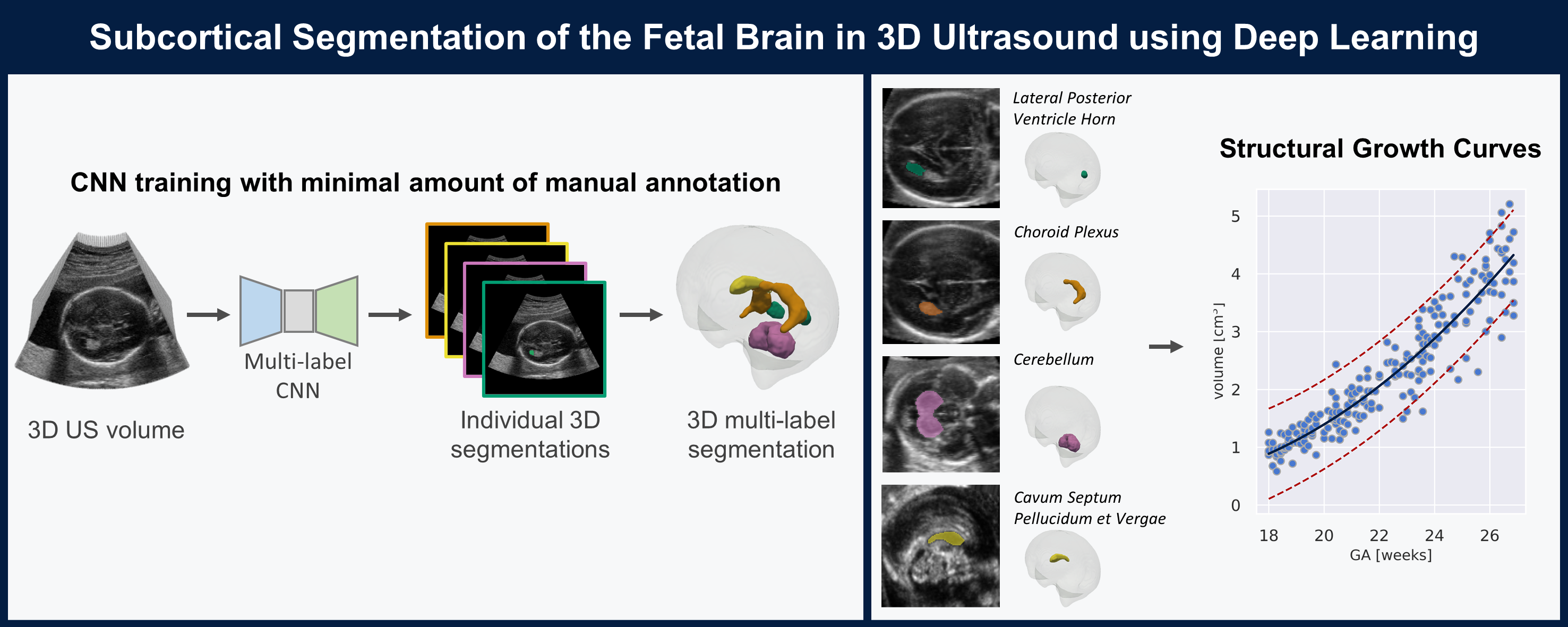
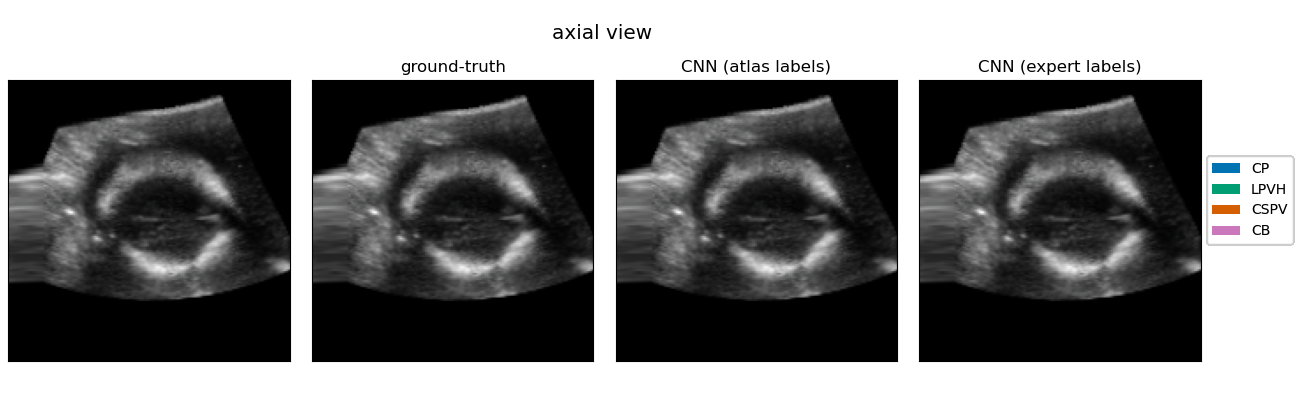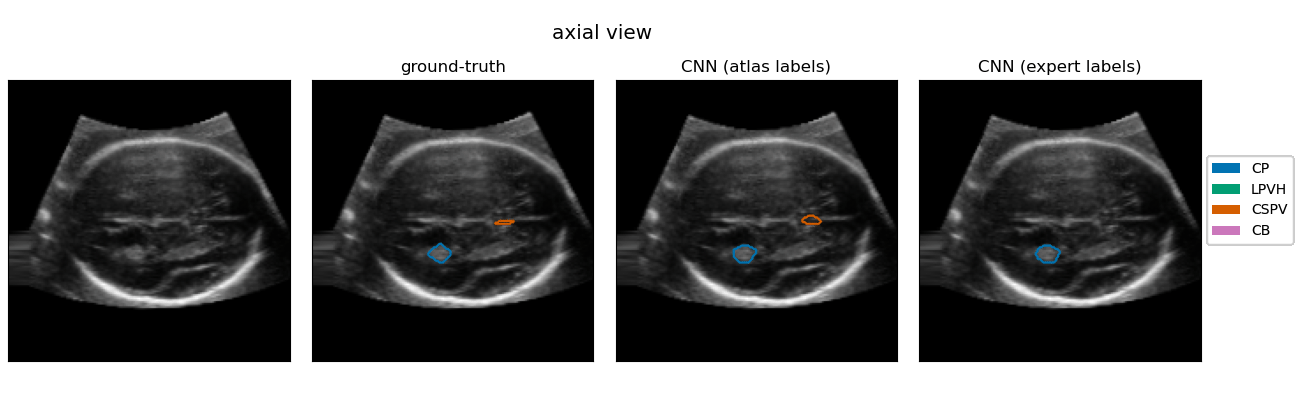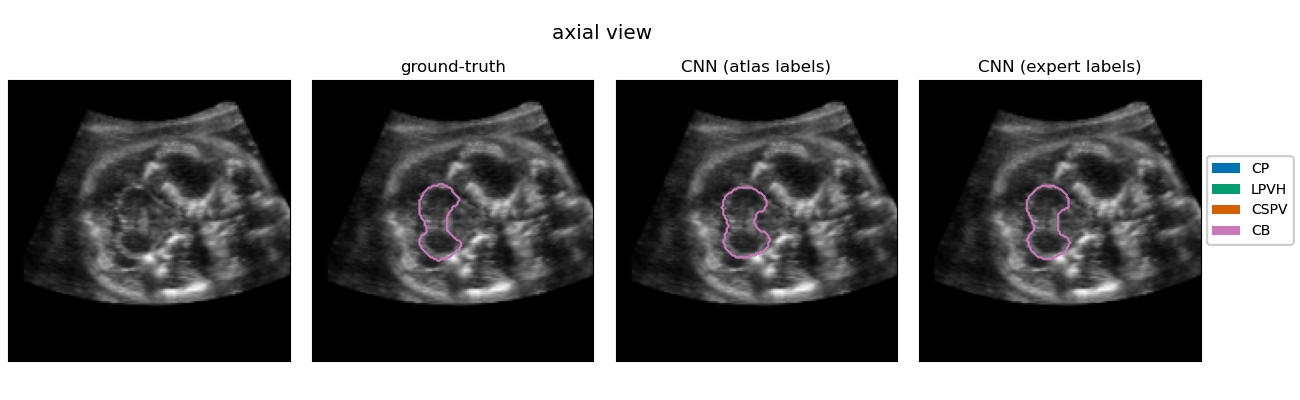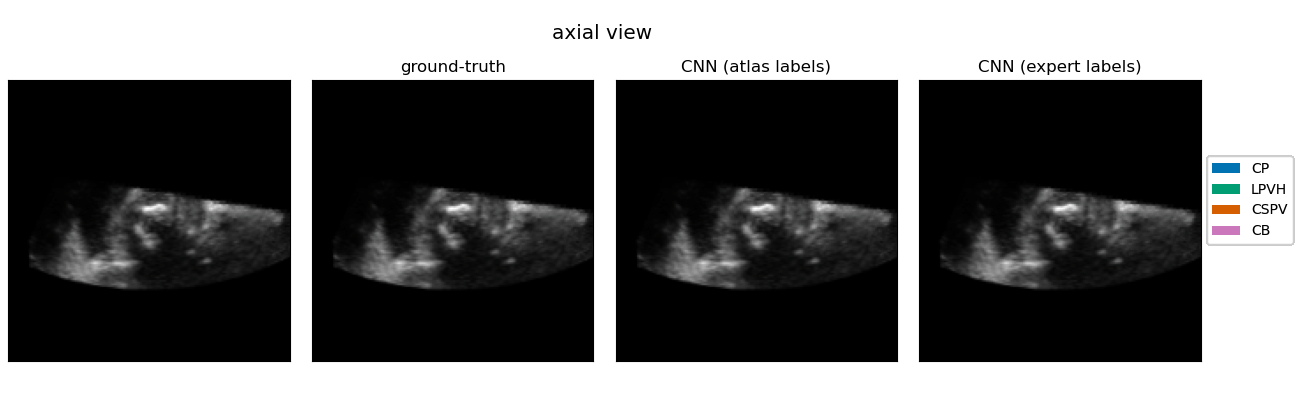
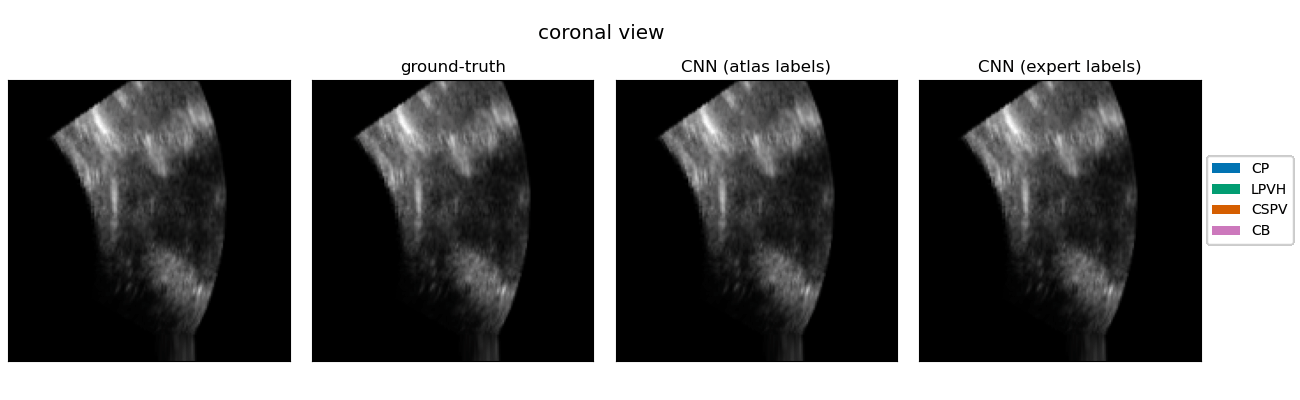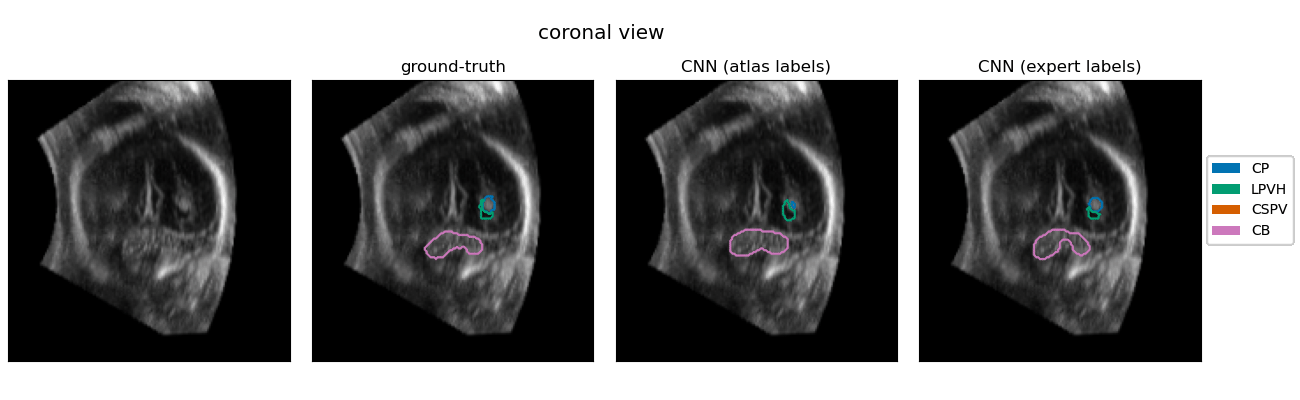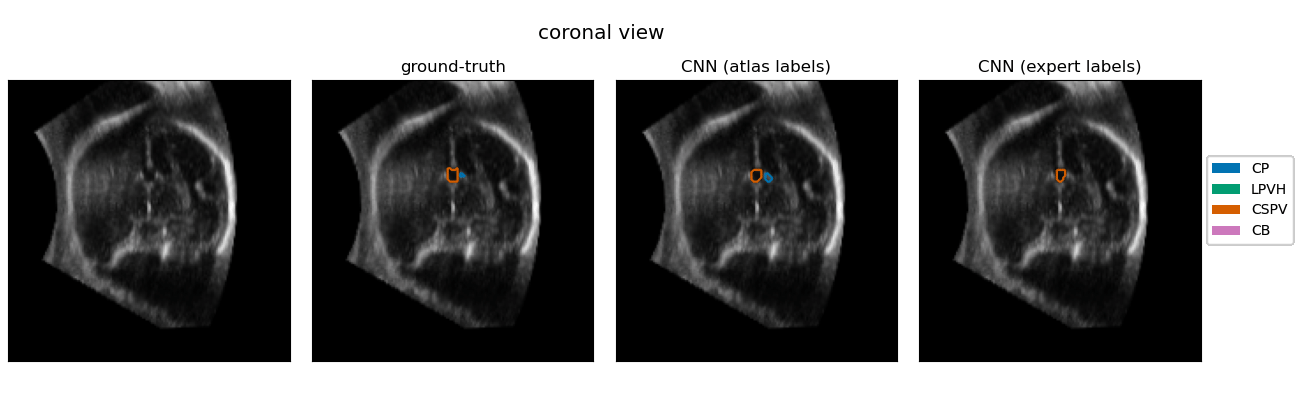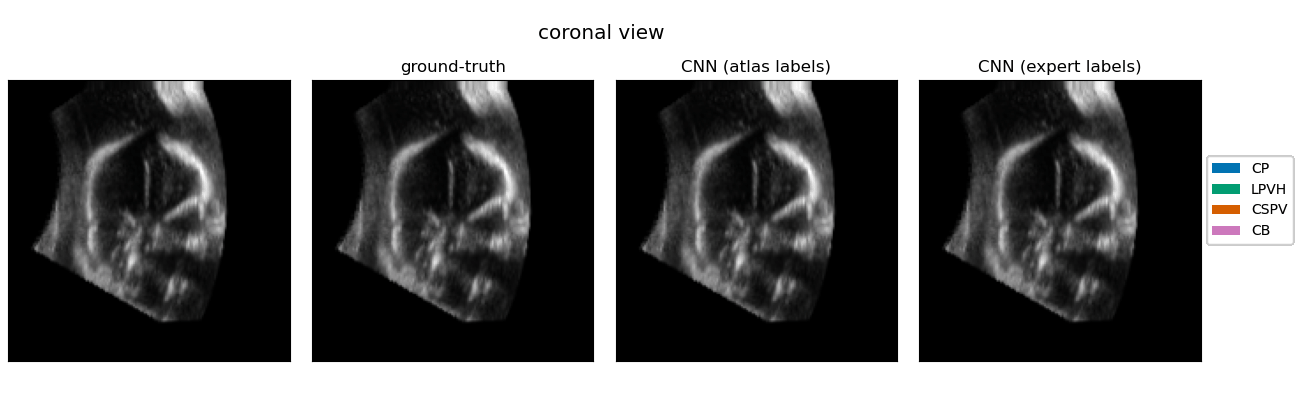
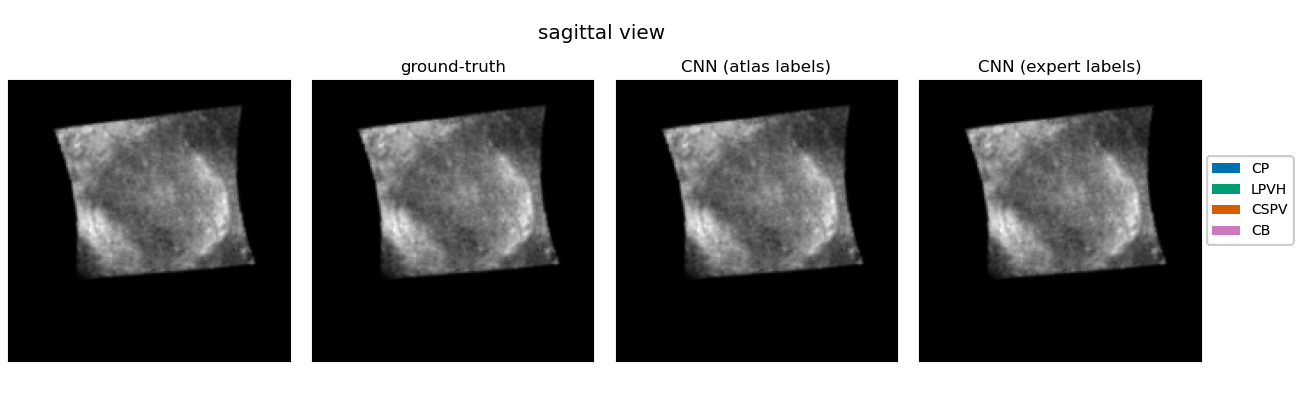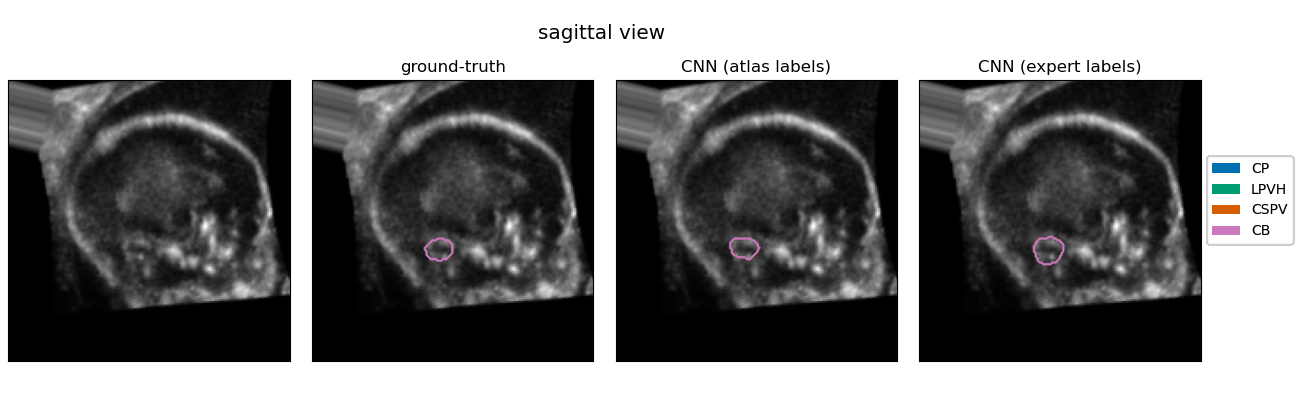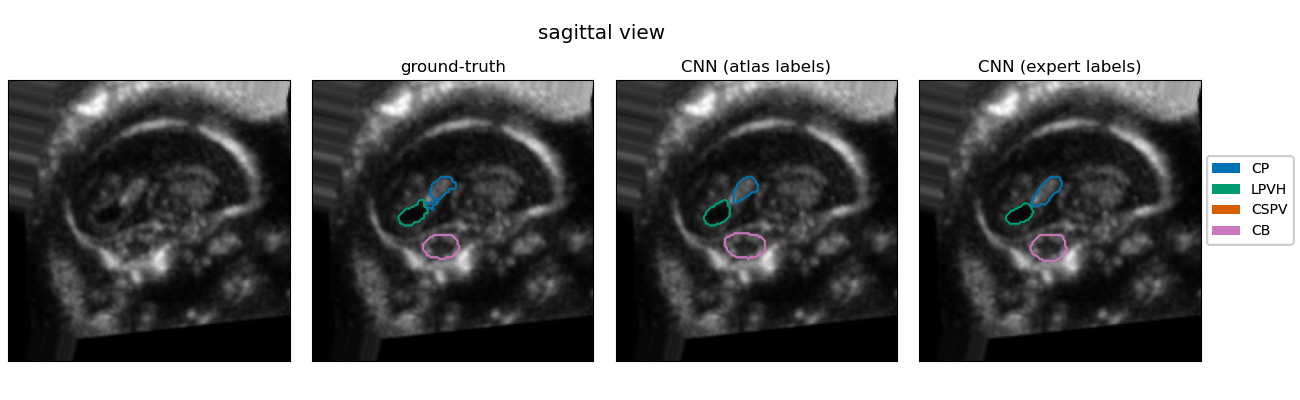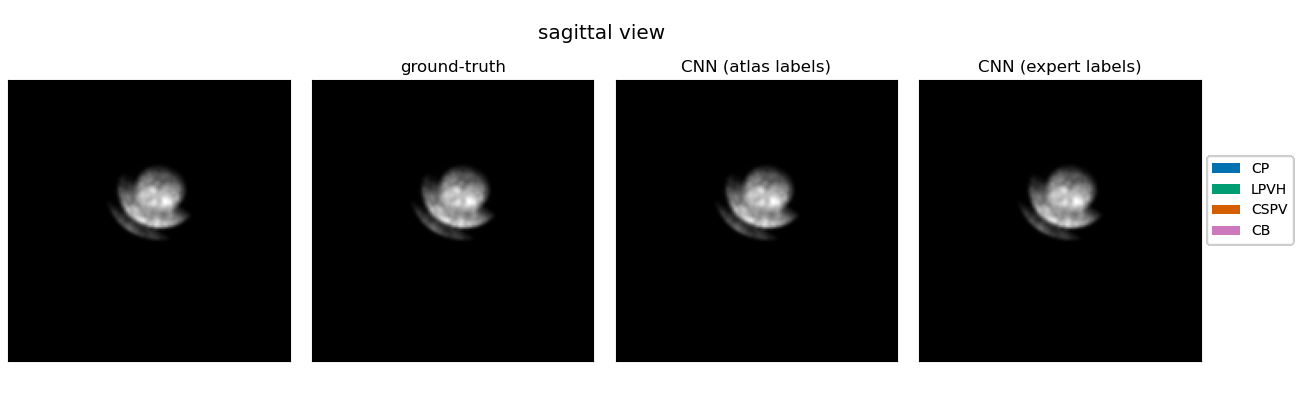
| The quantification of subcortical volume development from 3D fetal ultrasound can provide important diagnostic information during pregnancy monitoring. However, manual segmentation of subcortical structures in ultrasound volumes is time-consuming and challenging due to low soft tissue contrast, speckle and shadowing artifacts. For this reason, we developed a convolutional neural network (CNN) for the automated segmentation of the choroid plexus (CP), lateral posterior ventricle horns (LPVH), cavum septum pellucidum et vergae (CSPV), and cerebellum (CB) from 3D ultrasound. As ground-truth labels are scarce and expensive to obtain, we applied few-shot learning, in which only a small number of manual annotations (n = 9) are used to train a CNN. We compared training a CNN with only a few individually annotated volumes versus many weakly labelled volumes obtained from atlas-based segmentations. This showed that segmentation performance close to intra-observer variability can be obtained with only a handful of manual annotations. Finally, the trained models were applied to a large number (n = 278) of ultrasound image volumes of a diverse, healthy population, obtaining novel US-specific growth curves of the respective structures during the second trimester of gestation. |
Segmentation Pipeline: |
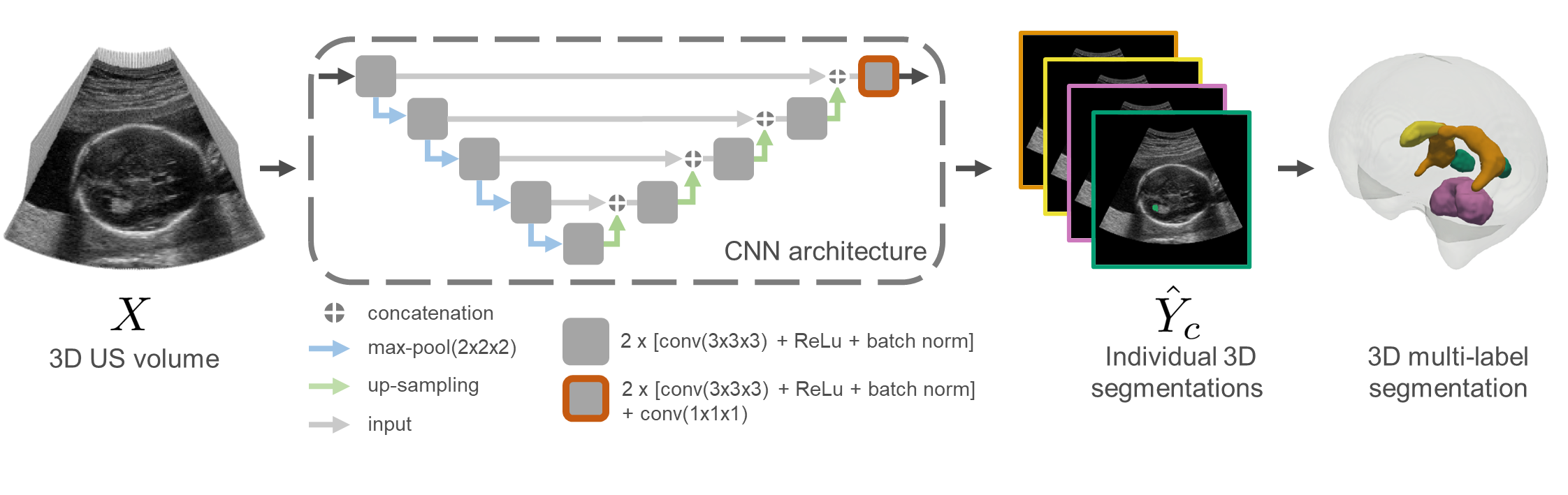
|
Atlas label generation |
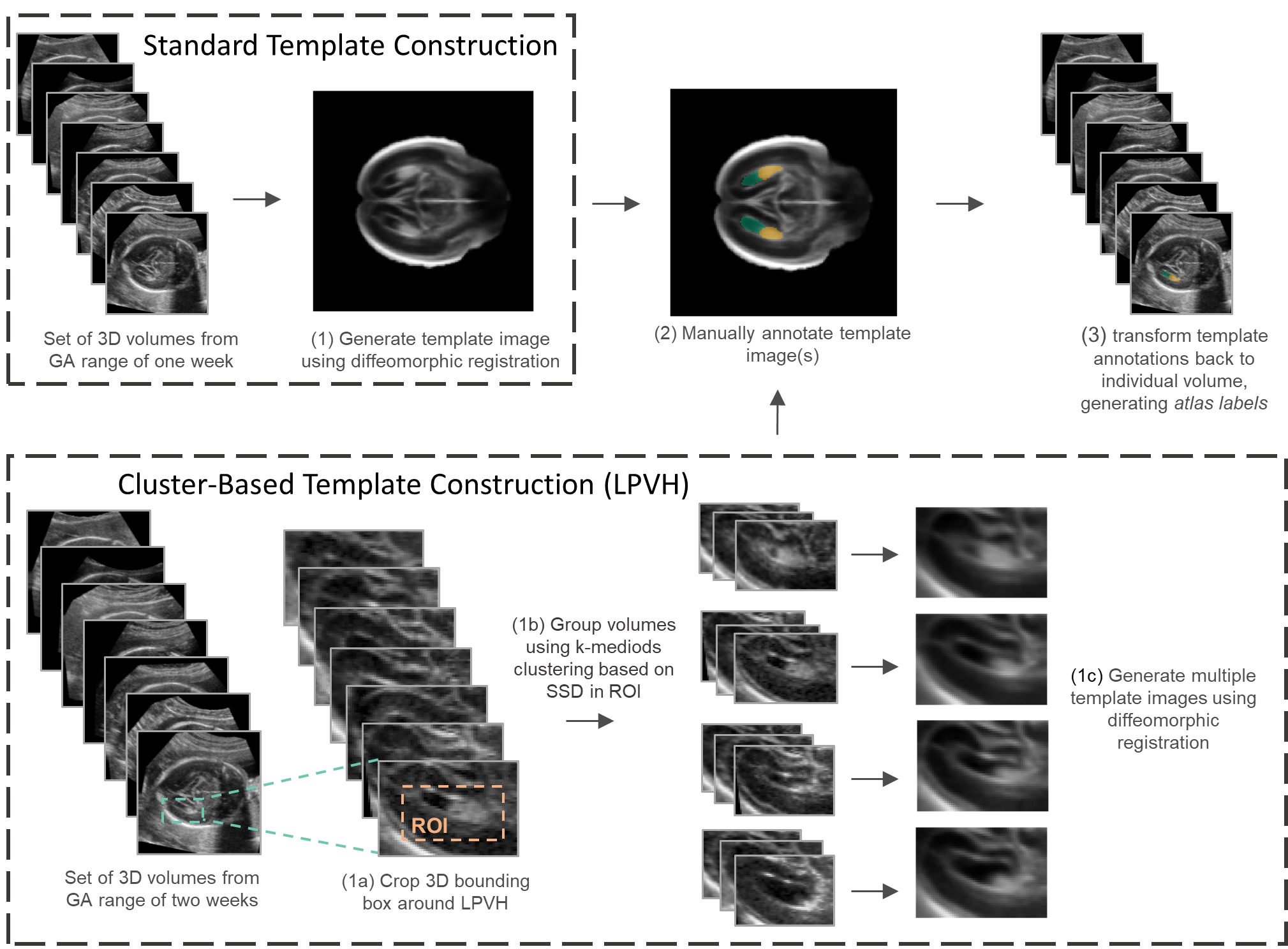
|
|
Schematic overview of atlas label generation. Standard whole brain templates (top row) were constructed for each GW and all structures (CB, CP, CSPV and LPVH) were annotated in these templates. Cluster-based template construction (bottom row) was only performed for the LPVH. |
|
|
|
|
|
|
|
|
|
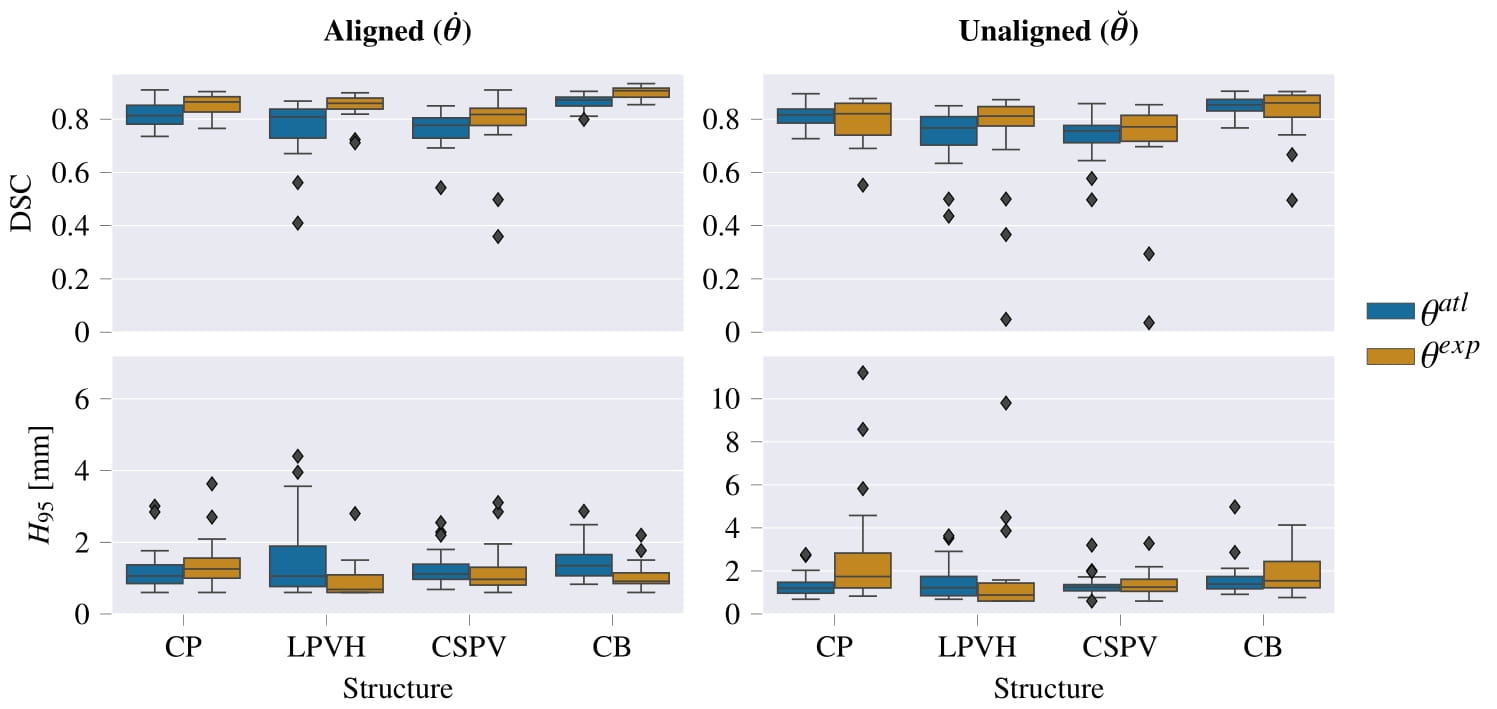
Resulting performance values after post-processing for a CNN trained with expert labels (θexp) and a CNN trained with atlas labels (θatl) |
|
|
|
|
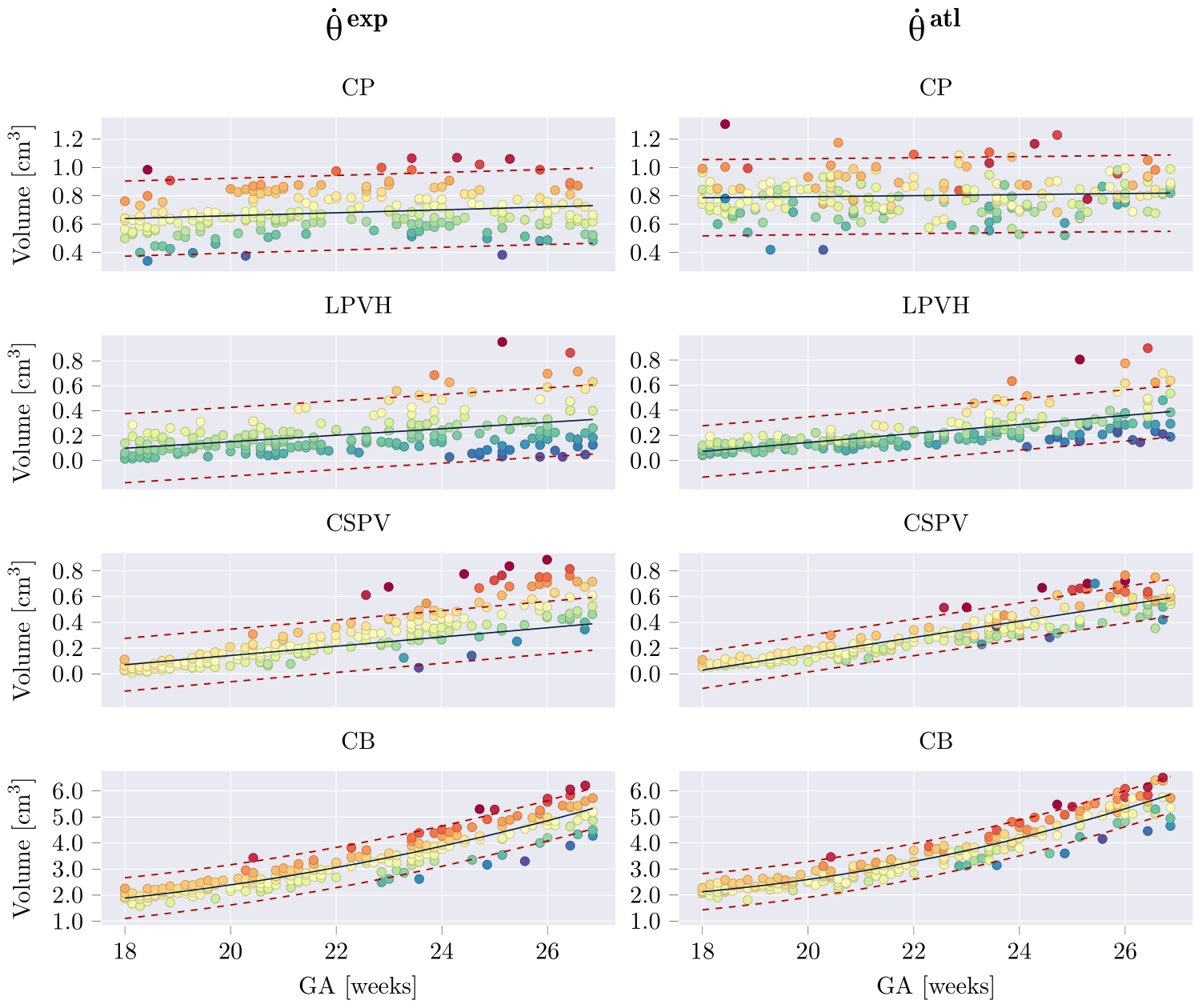
Estimated structural volumes for subcortical structures as a function of GA. Volumes were fitted with a linear or quadratic fit (black), in which the quadratic term was only added if it was significant for both networks (per structure). The 95% prediction confidence intervals where also computed and are shown with red dashed lines. For each structure, samples were colored based on their residual for θexp, and the same colors (per sample) where used for θatl. Relative volume predictions can be found in the paper. |
AcknowledgementsThis template was originally made by Phillip Isola and Richard Zhang for a colorful ECCV project; the code can be found here. |